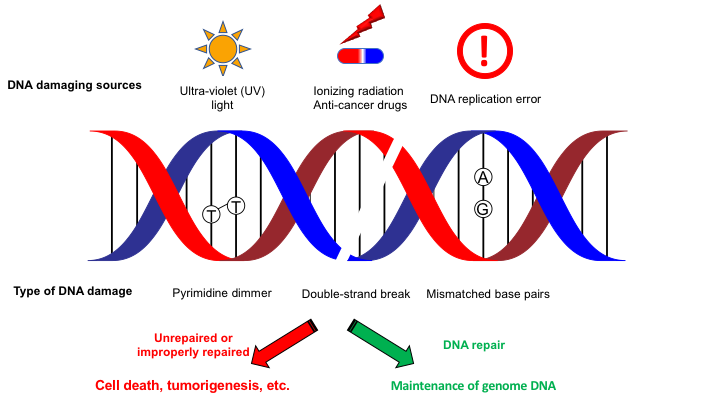
Genomic DNA that carries genetic information and should be maintained in safe is constantly challenged by various kinds of DNA damaging sources such as ultra-violet light and DNA replication error (Fig. 1). These DNA lesions that chemically differ can be repaired/removed by appropriate DNA repair machineries. If generated DNA lesions were not repaired or repaired in improper manner, DNA lesions can be causative of cell death and tumorigenesis, highlighting the biological importance of DNA repair mechanisms for maintaining genome integrity.
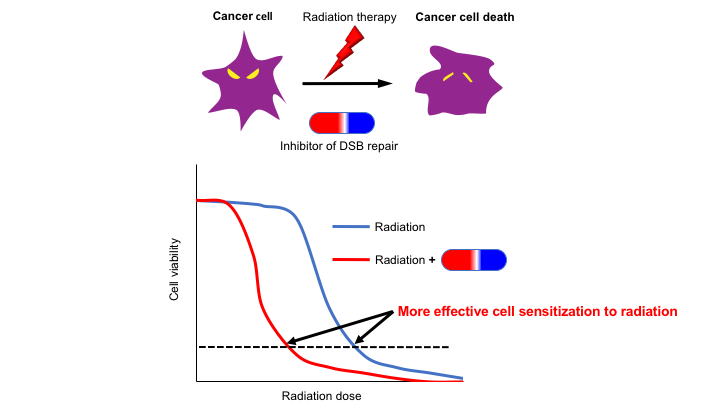
We aim to reveal the molecular mechanism of repairing DNA double-strand breaks (DSBs) that are generated by ionizing radiation (IR) and certain type of anti-cancer drugs in mammalian cells. DSBs that are formed by ablating both DNA backbones are one of the most deleterious type of DNA damages. Thus, understanding the molecular mechanism of DSBs generated by IR and anti-cancer drugs would make it possible to inhibit DSB repair only in cancer cells but not in normal cells, which will improve current anti-cancer therapy (Fig. 2). In addition, it is well known that defective DSB repair causes hereditary genetic disorders, which are characterized by a predisposition to cancer in many cases. Therefore, mechanisms of tumorigenesis caused by DSB repair defects also can be revealed by studying DSB repair.
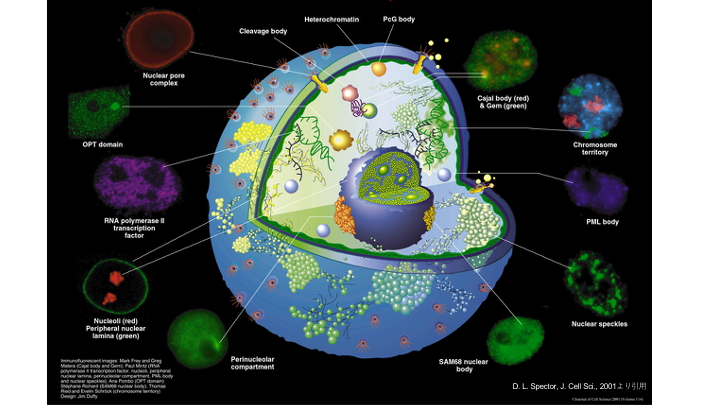
Research topic 1: Revealing crosstalk between nuclear bodies and DSB repair.
The basic molecular mechanisms of DSB repair have been extensively studies by many researchers including us, revealing how each DSB repair proteins function at the level of chromatin. On the other hand, nucleus is composed of various nuclear bodies (or nuclear structures) such as nucleoli, PML body and nuclear speckles in addition to genomic DNA. Although these nuclear bodies have been suggested to regulate distribution of genomic DNA and expression of genes, their involvement in DSB repair (and also in DNA repair) have not been actively studied (Fig. 3). Among these nuclear bodies, we are currently interested in how nuclear speckles that are involved in transcription and mRNA splicing can contribute DSB repair.
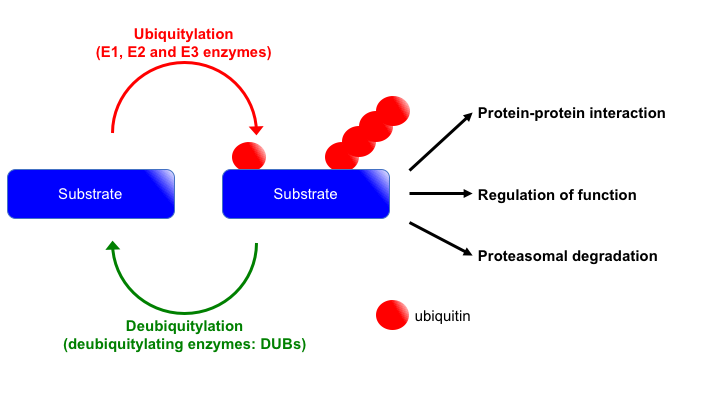
Research topic 2: Regulatory mechanisms of DSB repair by protein ubiquitylation
In cells, protein functions are regulated in various ways represented by post-translational modifications (PTMs) that include phosphorylation, acetylation and ubiquitylation. These PTMs play pivotal roles in many cellular functions including DSB repair. Importantly, these PTMs are reversible reactions. Namely, proteins can be activated, inactivated and/or recruited to specific locus by adding or removing these PTMs. Among these PTMs, we are studying how ubiquitylation regulates DSB repair. Although initially ubiquitylation was found as a signalling to lead proteins for proteasomal degradation, now it is well known that ubiquitylation plays key roles as a part of signal transduction by regulating protein-protein interaction and activity of proteins. As above mentioned, ubiquitylation is a reversible reaction. While addition of ubiquitin (mono, multi, or poly-ubiquitylation) is carried out by sequential reactions of E1, E2 and E3 enzymes, removing ubiquitin from proteins are performed by deubiquitylating enzymes (DUBs) (Fig. 4). To accomplish proper DSB repair, ubiquitylation and deubiquitylation must be orchestrated well. We are trying to understand ubiquitylation-mediated DSB repair regulation by identifying and characterizing DUBs involved in DSB repair.